Methodology
An Approach Published Since 2009
Currently, fragment-based topological design is a methodology that enables the creation of totally new molecules with desired biological profiles (potent activity, low toxicity, and adequate ADME properties). This methodology was created as a necessity due to the lack of “straightforward predictions” and too much money and time spent on discovering active and safe molecules. Fragment-based topological design focuses on simultaneously integrating several types of biological and chemical data into a single in silico model to create desirably new active and at the same time safe molecules, which can become, in almost no time, a new drug, pesticide, nanotherapeutic, or beauty product.
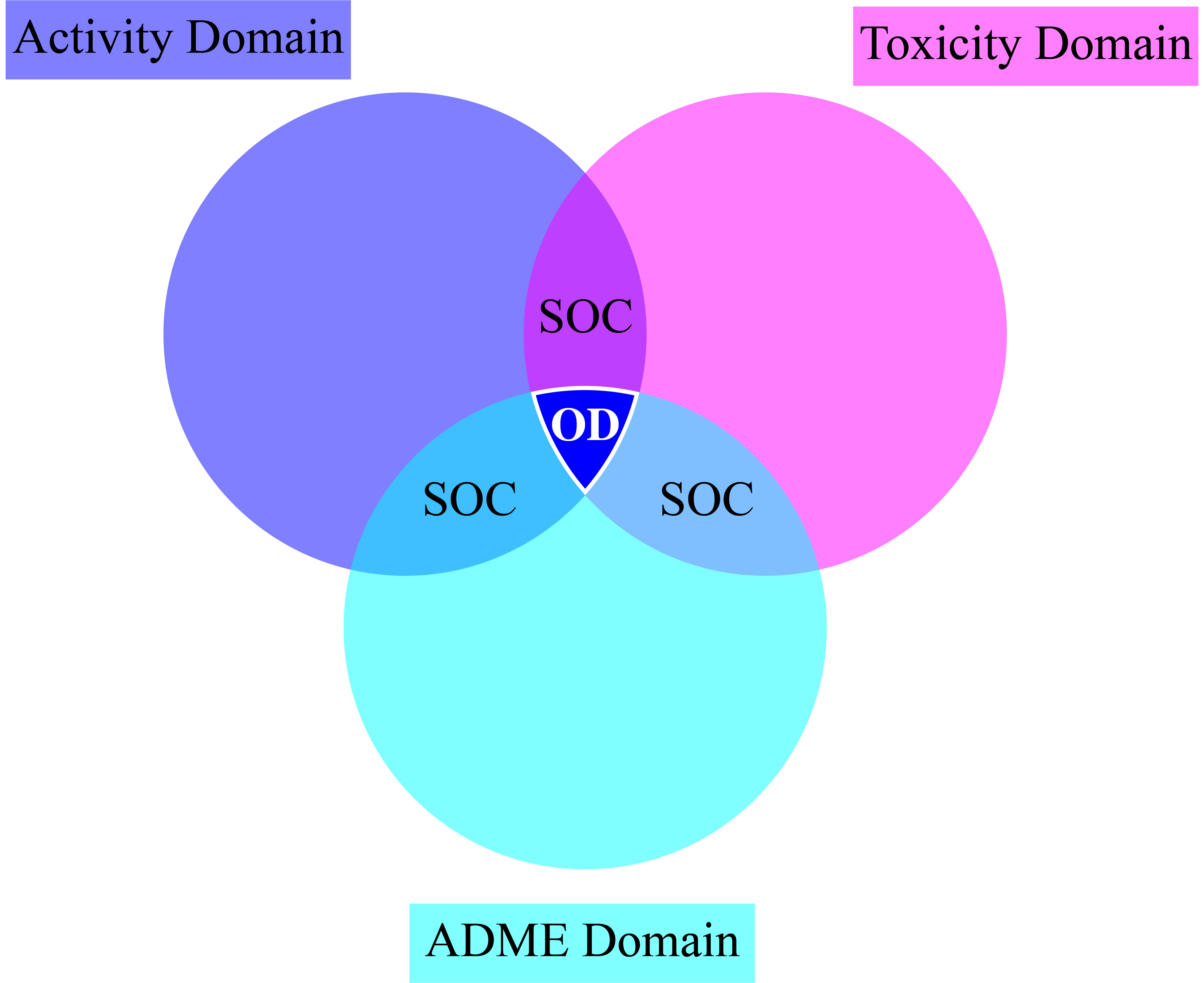
The OD (optimum drug) region is the zone to be targeted in drug discovery.
In other zones, we can find sub-optimal chemicals (SOC), which are formed by the overlapping of only two of the domains.
Beyond the SOC regions, molecules will tend to be discarded.
How could we target the OD region?
An emerging paradigm may hold the answer: fragment-based topological design
Speeding up Early Drug Discovery in Antiviral Research: A Fragment-Based in Silico Approach for the Design of Virtual Anti-Hepatitis C Leads
In this work, we report the first multitasking model for quantitative structure-biological effect relationships (mtk-QSBER), focused on the simultaneous exploration of anti-HCV activity and in vitro safety profiles related to the absorption, distribution, metabolism, elimination, and toxicity (ADMET). The mtk-QSBER model was created from a data set formed by 40 158 cases, displaying accuracy higher than 95% in both training and prediction (test) sets. Several molecular fragments were selected, and their quantitative contributions to anti-HCV activity and ADMET profiles were calculated. By combining the analysis of the fragments with positive contributions and the physicochemical meanings of the different molecular descriptors in the mtk-QSBER, six new molecules were designed. These new molecules were predicted to exhibit potent anti-HCV activity and desirable in vitro ADMET properties. In addition, the designed molecules have good druglikeness according to the Lipinski’s rule of five and its variants.

De novo computational design of compounds virtually displaying potent antibacterial activity and desirable in vitro ADMET profiles
In this work, we introduce the first multitasking model for quantitative structure-biological effect relationships focused on the simultaneous exploration of antibacterial activity against Gram-negative pathogens and in vitro safety profiles related to absorption, distribution, metabolism, elimination, and toxicity (ADMET). The multitasking model for quantitative structure-biological effect relationships was created from a data set containing 46,229 cases, and it exhibited accuracy higher than 97% in both training and prediction (test) sets. Several molecular fragments present in the compounds of the data set were selected, and their contributions to multiple biological effects were calculated, providing useful insights toward the detection of 2D pharmacophores, toxicophores, etc. Here, we used a fragment-based philosophy known as puzzle approach, where different fragments with positive contributions against all the biological effects (antibacterial activity and ADMET properties) were assembled as pieces of a puzzle, leading to the creation of six new molecules. Such assembly was dictated by the physicochemical interpretations of the different molecular descriptors of the model. The new molecules were predicted to exhibit potent activity against Gram-negative bacteria, and desirable ADMET properties. The druglikeness of these new molecules was in agreement with the Lipinski’s rule of five, making them promising candidates for future biological testing in the framework of collaborative drug discovery.